Related article: Detecting bacteria

Table of Contents
Microbes in the human body
According to a recent National Institutes of Health (NIH) estimate, 90% of cells in the human body are bacterial, fungal, or otherwise non-human.1) Although many have concluded that bacteria surely enjoy a commensal relationship with their human hosts, only a fraction of the human microbiotaThe bacterial community in the human body. Many species in the microbiota contribute to the development of chronic disease. has been characterized, much less identified. The sheer number of non-human genes represented by the human microbiota – there are millions in our “extended genome”2) compared to the nearly 23,000 in the human genome – implies we have just begun to fathom the full extent to which bacteria work to facilitate their own survival. 3)
The NIH's ongoing initiative, the Human MicrobiomeThe bacterial community in the human body. Many species in the microbiota contribute to the development of chronic disease. Project, aspires to catalog the human microbiome, also referred to as the human metagenome. Emerging insights from environmental sampling studies have shown, for example, that in vitroA technique of performing a given procedure in a controlled environment outside of a living organism - usually a laboratory. based methods for culturing bacteria have drastically underrepresented the size and diversity of bacterial populations. One environmental sample of human hands found 100 times more species than had previously been detected using purely culture-based methods. Another study which also employed high throughput genomic sequencing discovered high numbers of hydrothermal vent eubacteria on prosthetic hip joints, a species once thought only to persist in the depths of the ocean.
Human microbiota
Recent research has demonstrated that the diversity, prevalence and persistence of bacteria has been consistently underestimated. Microbes form most of the world's biomass: there are typically 40 million bacterial cells in a gram of soil and a million bacterial cells in a milliliter (gram) of fresh water.4) Studies have found bacteria in areas previously thought to be completely sterile. A broad diversity of bacteria were found at all of the “clean rooms” where NASA spacecraft are assembled and in spite of the highly desiccated, nutrient-bare conditions within.5)
Bacteria are no less persistent or proliferative inside the human body.
One prominent researcher called human skin a “virtual zoo of bacteria.”6) Another compared the diversity in the human gut to a rain forest.7) The human gut alone contains on average: 40,000 bacterial species,8) 9 million unique bacterial genes and 100 trillion microbial cells.9) According to Asher Mullard, “Between them [the bacteria in our bodies], they harbor millions of genes, compared with the paltry 20,000 estimated in the human genome. To say that you are outnumbered is a massive understatement.”10)
The global initiative known as the Human Microbiome Project currently estimates that the microorganisms that live inside or on Homo sapiens outnumber somatic (body) and germ cells [germ cells as in gametes, not bacteria] by a factor of ten.11) To this point, only approximately 1% of this microbiotaThe bacterial community which causes chronic diseases - one which almost certainly includes multiple species and bacterial forms. has been characterized and identified.12) The Human Microbiome Project aims to catalog the balance using an array of molecular sequencing techniques over the coming years.13) The combined genetic contributions of these microbes — in excess of 1,000,000 protein-coding genes — provide traits not encoded in our own genomes.14)
Since the inception of the Human Microbiome Project in 2007, dozens of research teams have gathered data which redefine what it means to be human. Some commentators have gone so far as to refer to the human body as a superorganism whose “whose metabolism represents an amalgamation of microbial and human attributes.”15)
Insights from new molecular methods for identifying bacteria
Researchers have long known that traditional methods for identifying bacteria are effective at identifying only a fraction of the bacteria in a given sample. New genomic based methods such as polymerase chain reaction (PCR) detect bacterial forms based on the presence of bacterial DNA or RNA. These new techniques are leading to some unexpected insights about bacteria.
- Bacteria are everywhere including the world's most hostile environments – According to Penn State researcher Jennifer Loveland-Curtze, “Microbes comprise up to one-third or more of the Earth’s biomass, yet fewer than 8,000 microbes have been described out of the approximately 3,000,000 that are presumed to exist,”
- NASA “clean rooms” – One would think that the the one place on Earth where bacteria do not exist is in the NASA “clean rooms” – the supposedly sterile places used to assemble aircraft. A 2007 research team compared the prevalence of bacteria found using traditional culture-based methods and ribosomal RNA gene sequence analysis. The four geographically diverse samples taken show a broad diversity in the types of bacteria able to grow in the most hostile environments including almost 100 types of bacteria, about 45 percent of which were previously unknown to science.16) The findings were something of a shock for NASA, an agency now forced to wonder exactly how many unknown pathogens have been taken to the moon and Mars.
- Two miles below the surface of a Greenland glacier – A Penn State team found viable “ultrasmall bacteria” in a glacial core17) – a habitat which is low-temperature, high-pressure, reduced-oxygen, and nutrient-poor. The core was estimated to be 120,000 years old.
- Deepest layer of the earth's crust – A 2010 analysis of the deepest layer of the Earth's oceanic crust has revealed a new ecosystem living over a kilometer beneath our feet. It is the first time that life has been found in the crust's deepest layer.18)

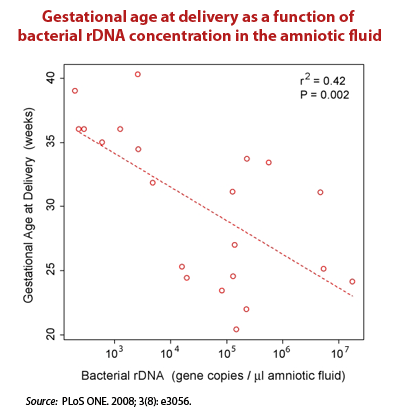
- In tissue sites once deemed sterile – The Relman Lab at Stanford used real time PCR to target conserved regions of the bacterial 16S ribosomal DNA (rDNA). They concluded that there is a substantial and “normal” population of bacterial DNA sequences in the blood of even healthy individuals.19)
- Antibiotic resistant strains predate human discovery of antibiotics – In a 2011 Nature study (press release), researchers carefully dug ancient 30,000 year old permafrost sediments out of the Canadian Northwest and sequenced the bacterial DNA found in it.20) The team concluded that antibiotic resistance genes predate our use of antibiotics and offers the first direct evidence that antibiotic resistance is an ancient, naturally occurring phenomenon widespread in the environment. This should not be surprising especially when one considers that penicillin came from a mold, tetracycline and demeclocycline from a strep mutant while Vancomycin came from Amycolatopsis orientalis.
- Each person has a unique mix of pathogens – A study led by Dr. Noah Fierer used a high-throughput method for PCR testing to identify the number and species of bacteria present on the hands of 51 undergraduate students leaving an exam room. Each student whose bacterial “fingerprint” – that is, their unique combination of bacteria – was sequenced, carried on average 3,200 bacteria from 150 species on their hands. Only five species were found on all the students’ hands, while any two hands – even belonging to the same person – had only 13% of their bacterial species in common.21)
- Communities of people have distinctive mixes of microbes – Two human ethnic groups based in India, which could not be distinguished on the basis of human DNA markers, could be distinguished based on their patterns of H. pylori variation.22)
- Communities of microbes associated with a disease may be less diverse than controls – irritable bowel syndrome,25) Crohn's disease,26) type I diabetes,27)AIDS28)
- Microbial variability may be relatively unrelated to food intake – While Dumas has shown that there are significant inter-regional differences in metabolites,29) work on the oral microbiome implies this may not be due to food consumption. In studying the oral microbiome, Nasidzie et al. took saliva samples from people in twelve diverse regions throughout the world including China, Germany, Poland, Congo, Philippines, and Louisiana.30) His team concluded that “while there is significantly more diversity in bacterial genera compared from different individuals than from the same individual, the diversity among individuals from the same location is nearly the same as the diversity among individuals from different locations.” The relative absence of variability between people in different regions implies that those factors which are highly variable, including food intake, may play a smaller than expected role in determining what at least a person's oral microbiota is. The Human Microbiome Research consortium concluded in 2012 that much of the diversity in healthy subjects remains unexplained.31)
- Many bacteria cannot be cultured using traditional cultivation techniques – Using PCR, Fierer's team found that the hands of student subjects contained 332,000 genetically distinct bacteria belonging to 4,742 different species. 45% of the species detected were considered rare. This marked a hundred-fold increase in the number of bacterial species detected in previous studies that had relied on purely culture-based methods (such as the Petri dish) to characterize the human hand microbiota.32) These conclusions are supported by the aformentioned study of NASA clean rooms, which found that only 0.1 to 55% of viable cells found via PCR were able to grow on defined culture medium.33)
- Some microbes need very few genes to persist – The genome of the microsporidia Encephalitozoon cuniculi (a pathogen which infects rabbits) is widely recognized as a model for extreme reduction and compaction. At only 2.9 Mbp, the genome encodes approximately 2,000 densely packed genes and little else. However, the nuclear genome of its sister, Encephalitozoon intestinalis, is even more reduced; at 2.3 Mbp, it represents a 20% reduction from an already severely compacted genome.34)
- A number of bacteria never thought to exist in man, do, and in large numbers. – A 2007 study, for example, found hydrothermal vent eubacteria on a prosthetic hip joint, which represents fully 6% of the bacteria sequenced and analyzed.35) Hydrothermal vent eubacteria otherwise grow best above 176°F (80°C).
- At least in fruit flies, gut microbes can alter mating preferences – In a 2010 study, mating preference was achieved by dividing a population of Drosophila melanogaster and rearing one part on a molasses medium and the other on a starch medium. When the isolated populations were mixed, “molasses flies” preferred to mate with other molasses flies and “starch flies” preferred to mate with other starch flies. The mating preference appeared after only one generation and was maintained for at least 37 generations. Antibiotic treatment abolished mating preference, suggesting that the fly microbiota was responsible for the phenomenon.36)
- Bacteria practice altruism – In a 2010 Nature paper, James J. Collins and his colleagues exposed one culture of Escherichia coli—some strains of which colonize the human and animal gut; others of which are notorious for causing disease outbreaks—to increasing amounts of an antibiotic over time. When they periodically analyzed the levels of drug resistance in the colony, they saw something unexpected: although the entire population was thriving in the presence of the drug, only a few individual bacteria were actually resistant. Further analysis revealed that the resistant mutants were secreting a molecule called indole that thwarts their own growth but helps the rest of the population survive by activating drug-export pumps on the bacterial cell membranes.37)
Forms of bacteria
Nobody can pretend to know the complete life cycle and all the varieties of even a single bacterial species. It would be an assumption to think so.
Ernst Almquist, a colleague of Louis Pasteur
Free-floating (planktonic) bacteria may be consistent with the popular conception of bacteria in the human body, but these types of bacteria are in the minority.38) Bacteria are distinguished by nothing if not their diversity – diversity in form, size, and habitat. Indeed, bacteria can float in the bloodstream, but they can also live inside human cells. They can exist in communities known as biofilm A structured community of microorganisms encapsulated within a self-developed protective matrix and living together.. One form of bacteria that has been studied for decades and about which a lot is known is the L-formDifficult-to-culture bacteria that lack a cell wall and are not detectable by traditional culturing processes. Sometimes referred to as cell wall deficient bacteria..
Bacteria regularly engage in “shape shifting” between forms. For example, Paenibacillus dendritiformis bacteria survive overcrowding by switching between two distinct vegetative phenotypes.39)
L-form bacteria
Main article: L-form bacteria
As a part of their natural life cycle, bacteria can transform into a variety of forms. One of those phases is the L-form.
L-form bacteria, also known as cell wall deficient bacteria, are a phase of bacteria that are very small and lack cell walls.
Though the subject of a great deal of research over the last 100 years and implicated in a variety of diseases, L-forms remain largely misunderstood - or at the very least, underappreciated - by the medical research community. According to the Marshall Pathogenesis, L-forms are part of a metagenomic microbiotaThe community of bacterial pathogens including those in an intracellular and biofilm state which cause chronic disease. responsible for chronic disease.
Biofilm bacteria
Main article: Biofilm bacteria
Biofilms are densely packed communities of microbial cells that grow on living or inert surfaces and surround themselves with secreted polymers. Many bacterial species form biofilms, and their study has revealed them to be complex and diverse. The structural and physiological complexity of biofilms has led to the idea that they are coordinated and cooperative groups, analogous to multicellular organisms.40)
Researchers have estimated that 60-80 percent of microbial infections in the body are caused by bacteria growing as a biofilm – as opposed to planktonic (free-floating) bacteria.
There is a perception that single-celled organisms are asocial, but that is misguided. When bacteria are under stress—which is the story of their lives—they team up and form this collective called a biofilm. If you look at naturally occurring biofilms, they have very complicated architecture. They are like cities with channels for nutrients to go in and waste to go out.
Andre Levchenko, PhD, Johns Hopkins University
Some external biofilm, namely chronic wounds and dental plaque, can be manually removed. Because of their inaccessibility and heightened resistance to certain antibiotic combinations and dosages, internal biofilm are more difficult to eradicate.
Biofilm bacteria are a part of what is known as the Th1 bacterial pathogens, which according to the Marshall Pathogenesis, collectively cause chronic disease. The Marshall Protocol targets the Th1 pathogens, in part, through the use of pulsed low doses of antibiotics, because they limit the growth of “persister cells.”
Biofilm-related disease. 41)
Other terms for very small bacteria
- Mollicutes - A class of bacteria distinguished by the absence of a cell wall. Emil Wirostko et al. found mollicute-like organisms in eyes of patients with sarcoidosis and Crohn's.42) In a later study, Wirostko was more specific, finding “mycoplasma-like organisms,” using the term the term Mycoplasma to the best-known genus of Mollicutes.43)
- Mycobacteria – A genus of Actinobacteria including tuberculosis and leprosy.
- nanobacteria – A proposed class of cell-walled microorganisms with a size much smaller than the generally accepted lower limit size for life (about 200 nanometers for bacteria). The existence of nanobacteria as organisms is debated. Some researchers argue that nanobacteria are actually calcifying nanoparticles.
Effects of bacteria on their human host
Main article: Effects of bacteria on their human host
Related articles: Acute infections, Acute respiratory infections
The genomes and the respective proteomes of microbes in the body frequently interact with those expressed by their human hosts. This is a key part of what is know as the interactome. The “massive”44) co-occurrence of protein-coding genes between microbes and humans speaks to the survival advantage of such homology, and the extent to which sequence overlap may play a key role in disease. Indeed, manipulation of host cell fate and orchestrated choreography of inflammatory responses are recurrent themes in the strategies of microbial pathogens.45) Bacteria affect host-cell pathways and human gene expression through a number of increasingly well-documented ways.
<html><!–

–></html>
It is what bacteria do rather than what they are that commands attention, since our interest centers in the host rather than in the parasite.
Theobald Smith, M.D., circa 1904 46)
2018 electron microscope study bacteria-in-your-brain
Successive infection and variability in disease
Main article: Successive infection and variability in disease
Related articles: Effects of bacteria and viruses on their human host, Familial aggregation
Chronic diseases manifest in patients and within patient populations with a high degree of variability. Some people have five chronic diseases, and others have one. Some patients experience symptoms of disease early in life while others not until they are very old. According to the Marshall Pathogenesis, this variability can be attributed to several factors.
Over the course of a lifetime, patients pick up the approximately 90 trillion bacteria to which they play host.47) While some researchers refer to each person's unique microbiota as an individual's “pathogen burden” and other terms,48) 49) we have referred to it as a person's “pea soupThe unique combination of bacterial pathogens (and co-mingling of bacterial genes) which accounts for each individual’s disease presentation..” In everyday language, the term pea soup is otherwise used to refer to a dense fog – an apt metaphor for the human microbiota. The promiscuity with which bacteria exchange DNA as well as the sheer number of bacteria to which any given person plays host are both factors which severely limit researchers' ability to accurately predict species-species and species-disease interactions.
The process by which a person accumulates the bacteria which drive disease is known as “successive infection.” In successive infection, an infectious cascade of pathogens slow the immune response and allow for subsequent infections to proliferate, resulting in dysbiosis (microbial imbalances). In patients sick with chronic inflammatory diseases, successive infection is ongoing and has additive properties: generally speaking, the more sick people are, the more sick they tend to become. Like a person's pea soup, the process by which a person accumulates additional bacteria via successive infection has an inherent variability to it.
Reconsidering classifying bacteria as species
Traditionally, bacteria have been understood to:50) 51)
- reproduce asexually
- not recombine their genetic material with other bacterial species
- be members of a clearly defined (or definable) species
- for a single species, be largely clones of one another
Recent analyses of bacterial DNA have revealed that these assumptions are misplaced. To a much greater extent than ever anticipated, bacteria rapidly and frequently share their DNA with their fellow prokaryotes – even distantly related bacteria – through a process called horizontal gene transfer.52) Other processes such as homologous recombination further muddle any kind of genomic coherence.53) As a result the diversity and variability among bacteria are much greater than anticipated.
Given the rapid diversification in the microbial world, it has become increasingly difficult to classify bacteria with traditional approaches.54) 55) When it comes to bacteria, the very definition of “species” may have to be reconsidered.56)
There's no single such thing as a microbial species. There's too much diversity in the range of biological collections that we might call species. Recognizing the variability between different groups, we'll probably abandon the notion of there being a single cutoff in terms of species definition…. The species concept is doomed to radical irrelevance because we don't actually need it any more. Metagenomics will come in and shift the paradigm for it…. More [novel] organisms are created through [genetic] recombination than through mutation.
W. Ford Doolittle, PhD speaking at Metagenomics 2006
For example, Hanage of Imperial College of London concluded that the classification of certain isolates of Neisseria was inherently “fuzzy.”57)

That said, there is some evidence that broad classifications of species appear more often in certain kinds of tissue:
Study of metagenomics
If species are defined by a shared gene pool, phylogenetic trees (such as the kind used to describe how Darwin's finches have common ancestors) do not satisfactorily model the relationships among bacteria – not when one organism could be a member of two or more otherwise quite distinct “species” simultaneously.60) One commentator suggests the relationship between bacteria is actually more like that of a web.61)
Enter metagenomics - a field which transcends the search for individual genomes. Literally “beyond genomics”62), metagenomics is an approach which looks at how whole communities of bacteria develop and interact including biofilm bacteria, intracellular bacteria, and L-form bacteria. Metagenomics provides a way of understanding the mysterious majority of microbes, which have been historically difficult to culture and classify. It is an approach, which involves taking a sample from the environment, pooling the DNA from all the different species present, fracturing it into a mixture of relatively short fragments and then sequencing the lot.
Metagenomics has begun to provide valuable insights into which communities of microbes cause disease. Given that each gene codes for a protein and that a number of proteins have harmful effects, the presence of a particular gene can and has signalled the presence of a pathogenic form of bacteria.
Metagenomic communities may cause disease
For more than a century, researchers have confined their thinking to Koch's Postulates, which erroneously dictated that a given infectious disease is always caused by a single microbial species. Indeed, a small minority of diseases such as leprosy are caused by a single pathogen.
However, over the years, researchers have cataloged ample evidence of why certain chronic diseases appear to be caused by pathogens: the inflammationThe complex biological response of vascular tissues to harmful stimuli such as pathogens or damaged cells. It is a protective attempt by the organism to remove the injurious stimuli as well as initiate the healing process for the tissue., the granuloma, the typical co-infections, the unique non-pathological microbial communities, etc. But, rarely have researchers found evidence of a single infectious agent, and that is because chronic diseases aren't caused by an individual species of microbe, but by ever-evolving, patient-specific whole communities of microbes. A fully realized understanding of metagenomics offers this key insight into chronic disease pathology.
The genomic diversity and relative importance of distinct genotypes within natural bacterial populations have remained largely unknown and may remain so for years to come.63)
The Marshall PathogenesisA description for how chronic inflammatory diseases originate and develop. makes no claims about which individual microbial species, if there are such things, are to blame for chronic disease. Besides, such a consideration is ancillary. The unique and difficult to define mix of pathogens an individual has is known as his or her pea soup – one of the definitions of which is “a dense fog.”
Role of Vitamin D Receptor
Main article: Metabolism of vitamin D and the Vitamin D Receptor
At least some of the bacteria which cause disease are intracellular. These microbes take hold progressively through a process called successive infectionAn infectious cascade of pathogens in which initial infectious agents slow the immune response and make it easier for subsequent infections to proliferate.. Chronic forms of bacteria are able to survive and reproduce by generating substances which block and turn off the Vitamin D ReceptorA nuclear receptor located throughout the body that plays a key role in the innate immune response., a key nuclear receptorIntracellular receptor proteins that bind to hydrophobic signal molecules (such as steroid and thyroid hormones) or intracellular metabolites and are thus activated to bind to specific DNA sequences which affects transcription. which controls the innate immune responseThe body's first line of defense against intracellular and other pathogens. According to the Marshall Pathogenesis the innate immune system becomes disabled as patients develop chronic disease.. So logical and powerful is this survival mechanism that it seems very likely that this is the primary mode by which chronic pathogenic forms persist. It simply makes evolutionary sense for pathogens to take full advantage of a receptor, which according to one recent study, transcribes hundreds of genes.64)
Acute infections
Main article: Acute infections
Related articles: Antibiotics under special circumstances, Effects of bacteria and viruses on their human host, Successive infection and variability in disease
The white blood cell count rises in cases of infection, steroid use and other conditions.
The immune system responds to cues in the microenvironment to make acute and chronic adaptations in response to inflammationThe complex biological response of vascular tissues to harmful stimuli such as pathogens or damaged cells. It is a protective attempt by the organism to remove the injurious stimuli as well as initiate the healing process for the tissue. and injury. The therapeutic significance of adenosine-mediated effects on the immune system is discussed here. 65)
The term acute infection is used to refer to microbe living inside a host for a limited period of time, typically less than six months. However, an abundance of research has emerged suggesting that acute infections have long-lasting effects, predisposing a person to later onset of chronic diseases.
The purpose of the Marshall ProtocolA curative medical treatment for chronic inflammatory disease. Based on the Marshall Pathogenesis. is to rehabilitate the immune response and improve the mix of microbes in the human body. In theory, this would free the immune response to target acute infections. Anecdotal reports from physicians and patients suggest that the MP is effective in this manner. To date, there have been no reports of tuberculosis or AIDS among MP patients.
Learn more
- Bacteria Provide New Insights Into Human Decision Making66) – “What each bacterium is doing is the equivalent if each individual on earth was able receive the exact information about the rate of spread of this new virus, the exact information about the intentions, to be vaccinated or not, by each person on the planet, and in addition the exact information about the health risks of side effects or being infected,” said Ben Jacob, the study's lead author. “A decision is then made in the context of this vast amount of information.”
- Darwin and microbiomes – Darwin did not mention microbes in his masterpiece, although Antoni van Leeuwenhoek had already reported their existence in the mid-seventeenth century; apparently, Darwin was not aware of this discovery. As Norman Pace commented in a recent talk, “On the Origin of Species was sterile, as it was not contaminated with bacteria.” Indeed, Darwin would have been astounded to know that some of the best evidence for natural selection resided in his own gastrointestinal tract.67)
- 'Social-IQ Score' for Bacteria Developed – discussion of a 2011 paper68) that developed a “Social-IQ score” for bacteria
- Exploring human microbiome research - Amy Proal 2015 blog MICROBE MINDED
[PMID: 17943116] [PMCID: 3709439] [DOI: 10.1038/nature06244]
[PMID: 20388071] [DOI: 10.2174/138161210790883840]
[PMID: 28245427]
[PMID: 9618454] [PMCID: 33863] [DOI: 10.1073/pnas.95.12.6578]
[PMID: 17655710] [DOI: 10.1111/j.1574-6941.2007.00360.x]
[PMID: 17293459] [PMCID: 1815283] [DOI: 10.1073/pnas.0607077104]
[PMID: 17014427] [DOI: 10.1111/j.1365-2222.2006.02579.x]
[PMID: 18043225] [DOI: 10.1097/MOG.0b013e3282f2b0e8]
[PMID: 19562079] [PMCID: 2699651] [DOI: 10.1371/journal.pone.0006074]
[PMID: 18509412] [DOI: 10.1038/453578a]
[PMID: 17620602] [PMCID: 1924555] [DOI: 10.1073/pnas.0704662104]
[PMID: 12037568] [DOI: 10.1038/417552a]
[PMID: 16741115] [PMCID: 3027896] [DOI: 10.1126/science.1124234]
[PMID: 17308177] [PMCID: 1855582] [DOI: 10.1128/AEM.03007-06]
[PMID: 16332755] [PMCID: 1317422] [DOI: 10.1128/AEM.71.12.7806-7818.2005]
[PMID: 21079766] [PMCID: 2974637] [DOI: 10.1371/journal.pone.0015399]
[PMID: 11326021] [PMCID: 88056] [DOI: 10.1128/JCM.39.5.1956-1959.2001]
[PMID: 21881561] [DOI: 10.1038/nature10388]
[PMID: 19004758] [PMCID: 2584711] [DOI: 10.1073/pnas.0807920105]
[PMID: 15051885] [PMCID: 387319] [DOI: 10.1073/pnas.0306629101]
[PMID: 21194740] [PMCID: 3037020] [DOI: 10.1016/j.jaci.2010.10.048]
[PMID: 18487399] [PMCID: 2519371] [DOI: 10.1128/AEM.02884-07]
[PMID: 19606407] [DOI: 10.1055/s-0028-1109055]
[PMID: 16188921] [PMCID: 1856500] [DOI: 10.1136/gut.2005.073817]
[PMID: 20613793] [PMCID: 3105672] [DOI: 10.1038/ismej.2010.92]
[PMID: 18691828] [DOI: 10.1016/j.mehy.2008.06.012]
[PMID: 16579598] [PMCID: 6561113] [DOI: 10.1021/ac0517085]
[PMID: 19251737] [PMCID: 2665782] [DOI: 10.1101/gr.084616.108]
[PMID: 22699609] [PMCID: 3564958] [DOI: 10.1038/nature11234]
[PMID: 20865802] [PMCID: 4355639] [DOI: 10.1038/ncomms1082]
[PMID: 17501992] [PMCID: 2206354] [DOI: 10.1186/ar2201]
[PMID: 21041648] [PMCID: 2993361] [DOI: 10.1073/pnas.1009906107]
[PMID: 20811456] [PMCID: 2936489] [DOI: 10.1038/nature09354]
[PMID: 11257008] [PMCID: 90417] [DOI: 10.1128/AAC.45.4.999-1007.2001]
[PMID: 21628502] [PMCID: 3104493] [DOI: 10.1128/mBio.00069-11]
[PMID: 19067751] [DOI: 10.1111/j.1574-6976.2008.00150.x]
[PMID: 29235402] [DOI: 10.1080/14787210.2018.1417036]
[PMID: 2801045] [DOI: 10.1111/j.1755-3768.1989.tb01626.x]
[PMID: 8140710] [PMCID: 1298461]
[PMID: 18582510] [PMCID: 7115663] [DOI: 10.1016/j.peptides.2008.05.022]
[PMID: 10845851] [DOI: 10.1161/01.atv.20.6.1417]
[PMID: 16138101] [DOI: 10.1038/nrmicro1236]
[PMID: 12446813] [DOI: 10.1093/oxfordjournals.molbev.a004046]
[PMID: 17255503] [PMCID: 2220085] [DOI: 10.1126/science.1127573]
[PMID: 11544367] [DOI: 10.1146/annurev.micro.55.1.561]
[PMID: 17020593] [PMCID: 1794555] [DOI: 10.1186/gb-2006-7-9-116]
[PMID: 15752428] [PMCID: 554772] [DOI: 10.1186/1741-7007-3-6]
[PMID: 17183309] [DOI: 10.1038/4441022a]
[PMID: 18502944] [PMCID: 2493393] [DOI: 10.1101/gr.075549.107]
[PMID: 16820057] [PMCID: 1560387] [DOI: 10.1186/1471-2164-7-171]
[PMID: 9818143] [DOI: 10.1016/s1074-5521(98)90108-9]
[PMID: 15731455] [DOI: 10.1126/science.1106028]
[PMID: 20736230] [PMCID: 2945184] [DOI: 10.1101/gr.107920.110]
[PMID: 20660309] [PMCID: 2922178] [DOI: 10.1073/pnas.1008254107]
[PMID: 19648955] [PMCID: 2726666] [DOI: 10.1038/embor.2009.166]
[PMID: 21167037] [PMCID: 3012674] [DOI: 10.1186/1471-2164-11-710]
[PMID: 20819230] [PMCID: 2996984] [DOI: 10.1186/1471-2164-11-488]
[PMID: 15843570] [DOI: 10.4049/jimmunol.174.9.5687]
[PMID: 12634792] [DOI: 10.1038/nature01510]